Introduction
Respiratory viral infections are commonly associated with secondary bacterial pneumonia. At the beginning of 2020, in Russia, secondary infections were identified in 59.55% of influenza A cases, 23.63% of influenza B cases, 21.26% of respiratory syncytial virus cases, and 42.42% of metapneumovirus cases.1 The frequency of bacterial infection in adult COVID-19 patients varies from 5 to 9% in reports from Europe to 45.5% in Thailand and 55.4% in the USA.2 The difficulty in differentiating a true infection from colonization may lead to overdiagnosis of bacterial infections. Taking into account this source of bias, up to 91.8% of COVID-19 patients may be considered to have bacterial co- or secondary infections, and up to 23.3% may be secondary infected by fungal pathogens,3,4 especially Candida spp.2,5
A series of preventive and control measures for SARS-CoV-2 during the COVID-19 pandemic contained the spread of SARS-CoV-2 and decreased the infection of other pathogens, including Mycoplasma pneumoniae and Chlamydophila pneumoniae,6 Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis.6–8 Therefore, in earlier reports, its frequency is low, with prevalence near 21% for bacterial and 12% for fungal co- or superinfections.9 Meanwhile, even at the beginning of the COVID-19 pandemic, in China, up to 80% of SARS-CoV-2-infected subjects had IgM positivity against at least one other respiratory agent.10 Following the reduction of pandemic restrictions, a resurgence of respiratory pathogen-spread cases appeared.7,8,11
Bacterial pneumonia is a common coinfection in COVID-19 patients, most frequently caused by H. influenzae, St. pneumoniae or Staphylococcus aureus. Other opportunistic pathogens, such as Klebsiella spp., Pseudomonas aeruginosa, Acinetobacter baumannii, and Escherichia coli, were also identified as predominant co- or super-infection causative agents in several studies.2,4,8,12–16 All these microorganisms may be community-acquired, persistent causative agents of an underlying chronic infection, or nosocomial.1,12 Such pathogens as St. pneumoniae and H. influenzae were found in the hospital aerosols.17 While St. pneumoniae remained the main causative agent of secondary pneumonia acquired out of hospital, S. aureus and H. influenzae often cause hospital-acquired pneumonia.1
Because bacterial coinfections and superinfections are widespread, empirical use of antibacterials specifically at the onset of the COVID-19 infection is justified.9 Coinfections or secondary infections in COVID-19 patients are associated with a poor prognosis.18 These patients also have a higher risk of admission to the ICU, septic shock, increased maximum CRP and ferritin values, higher proportions with fever, purulent sputum, peripheral leukocytosis, and consolidative opacity, and a longer duration of therapy.1–4,12,16,19,20
Meanwhile, it is suggested that bacterial infections may play a less relevant role in non-severe COVID-19.21 In the present study, we estimate the diversity of bacterial and fungal respiratory pathogens in patients with moderate forms of COVID-19 who were hospitalized but did not need ICU (except four patients) or mechanical ventilation. The purpose of this research was to evaluate the impact of co-infection on the severity of COVID-19, particularly in vaccinated individuals who are expected to be protected against the coronavirus infection.
Materials and Methods
Data collection
We retrospectively analyzed the clinical, immunological, and microbiological findings in patients with COVID-19 (n = 271), including COVID-19 and Ch. pneumoniae coinfection (n = 81), and COVID-19 and M. pneumoniae (n = 72) coinfection treated in the State Budgetary Institution of Healthcare in the Leningrad Region “Luga Interdistrict Hospital” in 2021–2022. In three subgroups, i.e., only COVID-19, COVID-19 and Ch. pneumoniae coinfection, and COVID-19 and M. pneumoniae coinfection, patients were selected randomly to reach the numbers 100, 100, and 100 subjects per group, respectively; however, part of the population was excluded from the analysis because of the lack of microbiological data.
All patients were examined at admission, and the severity of the lung involvement was estimated on the computed tomography (CT). The CT score was assessed as 0 for < 5% involvement, 1 for 5%-25% involvement, 2 for 26%-49% involvement, 3 for 50%-75% involvement, and 4 for > 75% involvement. The National Early Warning Score (NEWS) was estimated for all patients at admission according to NEWS2 standards.22 Data for CT score, NEWS, hospitalization duration, ICU, and previous anti-SARS-CoV-2 vaccination were collected from medical charts. None of the patients had Mycobacterium tuberculosis DNA in sputum.
Data for age, sex, sputum culture, anti-Ch. pneumoniae IgM, and anti-M. pneumoniae IgM were collected in the Laboratory Information System (LIS) of the North-West Centre for Evidence-Based Medicine laboratory as part of the routine diagnostic workflow and then re-obtained for the present study. All data were anonymized for the analysis.
All procedures performed in the study were in accordance with the ethical standards of the institutional research committee and national standards. For this type of retrospective study, formal consent is not required.
Microbial Isolation and Identification
The matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS) method was used for the identification of microorganisms in sputum. Clinical samples were cultivated in Columbia blood agar for 24 hours and Sabouraud agar, followed by colony transferring to the sample plate, matrix adding, and drying. MALDI Microflex Biotyper (Bruker Daltonics, Bremen, Germany) was used with α-cyano-4-hydroxycinnamic acid matrix according to the manufacturer’s guidelines.23
Following national guidelines for laboratory diagnosis of community-acquired pneumonia, MUK 4.2.3115-13,24 samples with 105 CFU/mL or higher are considered positive.
Automated ELISA for anti-Mycoplasma pneumoniae and anti-Chlamydophila pneumoniae IgM evaluation
At admission, blood samples were collected to estimate anti-M. pneumoniae and anti-Ch. pneumoniae IgM levels. Blood samples were studied in the North-West Centre for Evidence-Based Medicine. ELISA test kits “CHORUS MYCOPLASMA PNEUMONIAE IgM” (DIESSE Diagnostica Senese, Italy) and “CHORUS CHLAMYDOPHILA PNEUMONIAE IgM” (DIESSE Diagnostica Senese, Italy) were applied to automatic testing with Chorus Trio (DIESSE Diagnostica Senese, Italy) according to the manufacturer’s guidelines. The samples with INDEX (ratio between the OD value of the sample and that of the cut-off) above 1.1 were considered positive.
Statistical Analysis
To estimate the impact of identified pathogens on CT score, NEWS value, and hospitalization duration, we evaluated the normality of values distribution using the Anderson–Darling test with the nortest R package (version 1.0-4). The test rejects the hypothesis of normality with a p-value less than 0.05. Since the hypothesis of normality was rejected, differences between groups were assessed for significance using the Mann-Whitney-Wilcoxon test. Mean value and standard deviation (SD) were also estimated. Data were visualized with the ggplot2 R package.25
The microbial co-occurrence was analyzed and visualized using cooccur v.1.3. R package.26 Only genera (or species) identified in at least five samples per group were analyzed.
Results
Study population
The study population included 271 COVID-19 patients, who were treated in the infectious department of the Luga Interdistrict Hospital. Although most patients were from the Luga district of the Leningrad region of Russia, some patients were from the nearby parts of the Leningrad region, especially the Gatchina district and Slantsy district, and Saint Petersburg. The details for CT score, NEWS value, duration of hospitalization, age, and sex distribution in the population are summarized in Table 1.
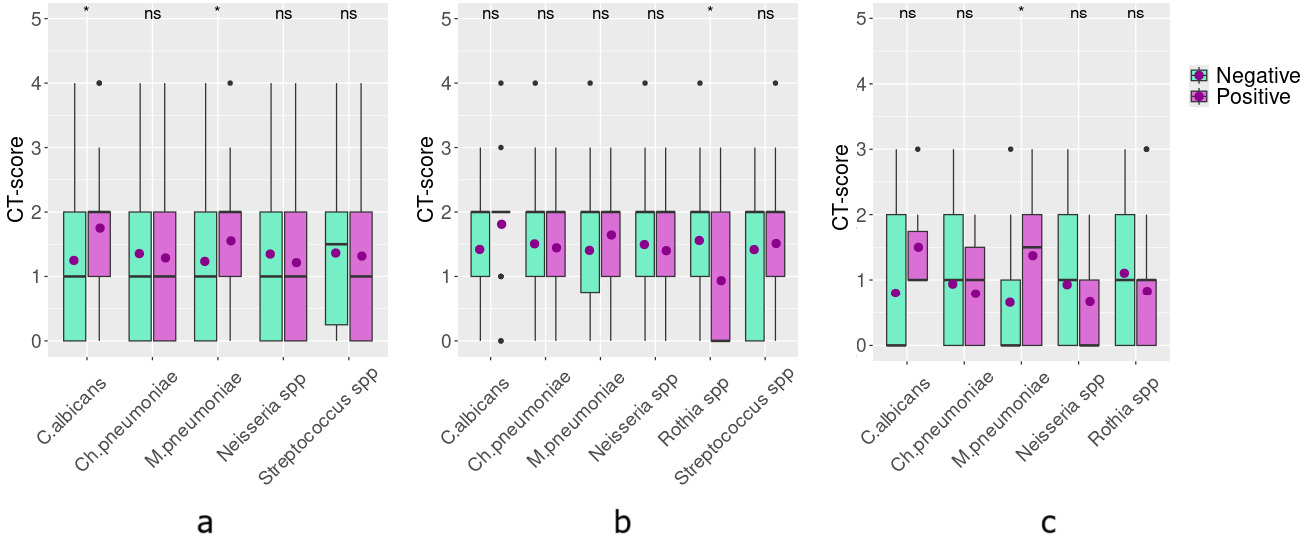
The CT score and NEWS values were favourable, and the duration of hospitalization was shorter in the vaccinated subjects (p < 0.05). Considering the potential differences in the disease progression in vaccinated and non-vaccinated patients, we further studied these groups separately. No differences in pathogen distribution were identified between vaccinated and unvaccinated groups (Figure 1a). In both groups, non-pneumococcal Streptococci were the predominant airway-colonizing bacteria (70% of the entire population; only one case of St. pneumoniae), followed by C. albicans (15.6% of the entire population) and Neisseria spp. (13% of the entire population).
When we evaluated the co-occurrence of different microorganisms in the population, we revealed the association between respiratory tract colonization by Neisseria spp. and Streptococcus spp., Neisseria spp. and Rothia spp. (including R. mucilaginosa, n = 36, and R. dentocariosa, n = 5), and Neisseria spp. and Haemophilus spp. (including H. influenzae, n = 1; H. parainfluenzae, n = 10; H. haemolyticus, n = 6; H. parahaemolyticus, n = 1), which was significant only in unvaccinated patients (Figure 1bc). Furthermore, M. pneumoniae coinfection was negatively associated with airway colonization by Rothia spp. The identified coexclusion between Ch. pneumoniae and M. pneumoniae may be associated with selection bias and should be interpreted with caution. Ch. pneumoniae and M. pneumoniae IgM-positive patients were revealed in both groups (nine subjects were unvaccinated and three were vaccinated).
Association between coinfections or airway colonization by opportunistic pathogens and disease severity on admission
Pathogens that were identified in less than 10% of the population were excluded from the analysis to avoid bias related to high group inequality. The association between C. albicans, Ch. pneumoniae, M. pneumoniae, and Streptococcus spp. infection or airway colonization with disease severity expressed in numerical form as CT score and NEWS was estimated. Because of the slightly higher frequency of Rothia spp. in unvaccinated patients (12%), we also evaluated its effect in this group.
In the entire population, lower respiratory tract colonization with C. albicans and anti-M. pneumoniae IgM positivity was significantly associated with higher CT scores, i.e., more pronounced lung damage (p < 0.05, Figure 2a). The association between anti-M. pneumoniae IgM positivity and lung damage is reproduced in vaccinated patients (p <0.05, Figure 2c), but does not occur in the unvaccinated group (Figure 2b). For C. albicans, the association with more pronounced lung damage lost the statistical significance when both subgroups were analyzed separately. Also, in unvaccinated patients, we identified a negative association between colonization by Rothia spp. and a higher CT score (p < 0.05).
No associations with worse NEWS values were identified for any pathogen included in the analysis, both in vaccinated and unvaccinated patients.
Association between coinfections or airway colonization by opportunistic pathogens and the duration of hospital stay
When we estimated the impact of M. pneumoniae, Ch. pneumoniae, and the most frequent (>10% of cases) airway-colonizing bacteria on the hospitalization duration in our study group, neither atypical pneumonia causative agents nor colonization with opportunistic pathogenic bacteria were associated with prolonged hospital duration in the studied patient group. Meanwhile, the patients in whom C. albicans was isolated from sputum stayed in hospital a bit longer than others (11.8±5.66 days vs. 13.6±6.07 days, p < 0.05, Figure 3a). However, this tendency did not reach statistical significance when the groups of vaccinated and non-vaccinated patients were analyzed separately (Figure 3bc).
Discussion
Commonly, the lung microbiome exhibits dynamic fluxes of microbial immigration and clearance. Thus, it demonstrates a fluid nature rather than a fixed one.28 SARS-CoV-2 damages airway epithelium, promotes inflammation, and deregulates immunity and interferon counteraction towards bacterial colonization and proliferation.2 Antimicrobial resistance of pathogens, aggressive use of immunomodulatory therapies, and overuse of antibiotics could contribute to the emergence of lung colonization by pathogens and secondary infections.13,21
In our study, we identified a high frequency (86.1%) of lung colonization by bacteria or fungi; however, it was mostly opportunistic pathogens or commensal bacteria, like Neisseria sp. Also, despite the dynamic nature of lung microbiota, we identified several trends in microbial co-occurrence and co-exclusion in unvaccinated patients, and some of them may have clinical significance, as discussed below.
It is suggested that, despite the high risks related to superinfections in severely ill COVID-19 patients, these infections play a less relevant role in the early, non-severe stages of COVID-19.29 In our data, we also did not find any correlations between patients’ condition (i.e., CT score and NEWS) and the majority of opportunistic pathogens isolated from sputum in follow-up. However, in our population, we revealed the association between suggested M. pneumoniae coinfection (i.e., anti-M. pneumoniae IgM positivity at hospital admission) in COVID-19 patients and the grade of lung damage on CT. This relation is significantly more evident in vaccinated patients. The symptoms of COVID-19 and M. pneumoniae-associated pneumonia are similar, with fever, cough, and shortness of breath, and these diseases demonstrate similar radiologic findings.30–32 Despite the importance and informativeness of CT in COVID-19 diagnosis, the sensitivity and specificity of pure ground-glass opacities are not very high, because many diseases similarly exhibit ground-glass opacities on chest imaging, including acute lupus pneumonia, diffuse interstitial pneumonia, pulmonary collagen vascular disease, acute eosinophilic pneumonia, and atypical pathogen infection in the lungs.33,34
M. pneumoniae infections are commonly comorbid with respiratory viral diseases.35 We suggest that in M. pneumoniae-infected anti-SARS-CoV-2-vaccinated COVID-19 patients, this atypical pathogen may have a significant impact on disease severity in a notable proportion of patients who are protected from COVID-19 by immunization. Under previously published data, coinfection with M. pneumoniae is associated with higher mortality, length of hospital stay, and the need for mechanical ventilation in COVID-19 patients.36–38 Meanwhile, in our population, we did not reveal any associations between M. pneumoniae coinfection and the duration of hospitalization, apparently because of successful empirical fluoroquinolone treatment and mild illness severity.
As was previously identified, approximately 10% of hospitalized COVID-19 patients have positive respiratory fungal cultures across multiple studies, and the Candida spp. is the most frequently isolated fungal pathogen.20 In patients from the Luga hospital, we identified a slight but significant impact of C. albicans on the duration of the disease. All patients received antibiotics from the first day of hospitalization; however, no antifungal treatment was administered prior to microbiological testing, and this is the most probable reason for the identified association. The positive association between an unfavourable CT score at hospital admission and a high C. albicans CFU number in sputum indicates that in the studied population, this pathogen may have originated from the community or patients’ commensal microflora rather than from a nosocomial environment.
Suddenly, we revealed the association between more favourable CT and Rothia spp. isolation from sputum in the follow-up. Previously, data demonstrated the controversial role of this opportunistic pathogen in COVID-19 patients. It was identified that great Rothia spp. abundance may be associated with higher lung damage or higher risk of hospitalization,5 but inversely associated with severe COVID-19.39 Apparently, Rothia spp does not develop severe illness in immunocompetent moderately ill COVID-19 patients who receive appropriate antibiotic therapy.40 Furthermore, in our study, we revealed coexclusive relationships between anti-M. pneumoniae IgM and isolation of Rothia spp. from sputum. As described above, anti-M. pneumoniae IgM-positive patients have more pronounced ground-glass opacities on CT. Therefore, it cannot be excluded that the negative association of Rothia spp. and lung damage may be prone to bias related to the selection mode.
Limitations
The present study’s major limitation is its focus on a narrow, homogeneous group of patients with moderately severe disease, all treated in the same department. The group of patients is relatively small and does not allow for estimating the impact of some pathogens on disease severity and hospitalization duration. Also, there is some disproportion in the patient selection. So, in our group, 31% of patients have anti-M. pneumoniae IgM and 35% of patients have anti-Ch. pneumoniae IgM, despite the actual proportion of such patients in Luga Hospital being 5.21% and 6.67%, respectively.27 Furthermore, the sputum samples were acquired for a microbiological study on different hospitalization days, which does not allow us to differentiate nosocomial infections from community-acquired cases. Similarly, it is difficult to distinguish a true infection from colonization, which may lead to an overdiagnosis of bacterial infections.18 especially in such samples as sputum, which is inevitably contaminated by oral flora. In the present study, we apply 105 CFU/mL as the detection limit for a positive sputum culture according to national guidelines for laboratory diagnosis of community-acquired pneumonia, MUK 4.2.3115-13 to discriminate clinically significant microbial growth.24
Despite these limitations, we confirm the level of pathogenic microorganism coinfection and superinfections is higher among COVID-19 patients, including subjects with moderate disease. Also, we revealed that coinfection, like M. pneumoniae, may significantly impact the disease severity in vaccinated patients. Meanwhile, the empirical use of antibacterial treatment offsets the negative impact of these pathogens in a moderate form of COVID-19 infection.
Conclusion
Considering the wide distribution of bacterial and fungal coinfections and superinfections in COVID-19 patients, we estimated the impact of the bacterial pathogens Ch. pneumoniae and M. pneumoniae and lung colonization with opportunistic pathogens from normal microbiota on the disease severity and duration in patients with moderate COVID-19 infection. We have revealed a negative effect of M. pneumoniae coinfection on lung appearance on CT; however, the general state didn’t get worse in patients coinfected with SARS-CoV-2 and M. pneumoniae. The identified lung damage may mirror specific features of M. pneumoniae-related disease rather than a serious health risk. In view that this association is more pronounced in vaccinated patients, it may be speculated that the coinfection with M. pneumoniae rather than COVID-19 may be the reason for pneumonia onset and hospitalization, at least in vaccinated patients. Another important issue is a slight but significant association of C. albicans coinfection and longer hospitalization duration. The studied patient group was empirically treated with antibiotics from the first day of hospitalization, including third-generation cephalosporins and macrolides or fluoroquinolones. The delayed antifungal medication may be the reason for the identified C. albicans colonization effect. However, the increase in disease duration was not too high, and C. albicans coinfection was not too frequent to assume that empiric antifungal treatment would be particularly beneficial in this population.
Funding
The authors acknowledge Saint-Petersburg State University research grant 129659216.
Conflict of Interest
All authors have completed the ICMJE uniform disclosure form and declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Data accessibility statement
All data underlying the results are available as part of the article, and no additional source data are required.
Ethical Approval
All procedures performed in the study were in accordance with the ethical standards of the institutional research committee and national standards. For this type of retrospective study, formal consent is not required.
AI Statement
The authors confirm that no generative AI or AI-assisted technology was used to generate content.

_(whiskers_represent_the_confidence_interval)_and_co-occurrence_of_different_.png)


_(whiskers_represent_the_confidence_interval)_and_co-occurrence_of_different_.png)